Home
BGCFlow Wrapper



A snakemake wrapper and utility tools for BGCFlow, a systematic workflow for the analysis of biosynthetic gene clusters across large genomic datasets.
For more details, see documentation.
Please refer to the BGCFlow WIKI for detailed examples and use cases:

Publication
Matin Nuhamunada, Omkar S Mohite, Patrick V Phaneuf, Bernhard O Palsson, Tilmann Weber, BGCFlow: systematic pangenome workflow for the analysis of biosynthetic gene clusters across large genomic datasets, Nucleic Acids Research, 2024;, gkae314, https://doi.org/10.1093/nar/gkae314
Setup
Setup via Conda
To install bgcflow_wrapper with conda/mamba, run this command in your
terminal:
| # create and activate new conda environment
mamba create -n bgcflow -c conda-forge python=3.11 pip openjdk -y
conda activate bgcflow
# install BGCFlow wrapper
pip install bgcflow_wrapper
|
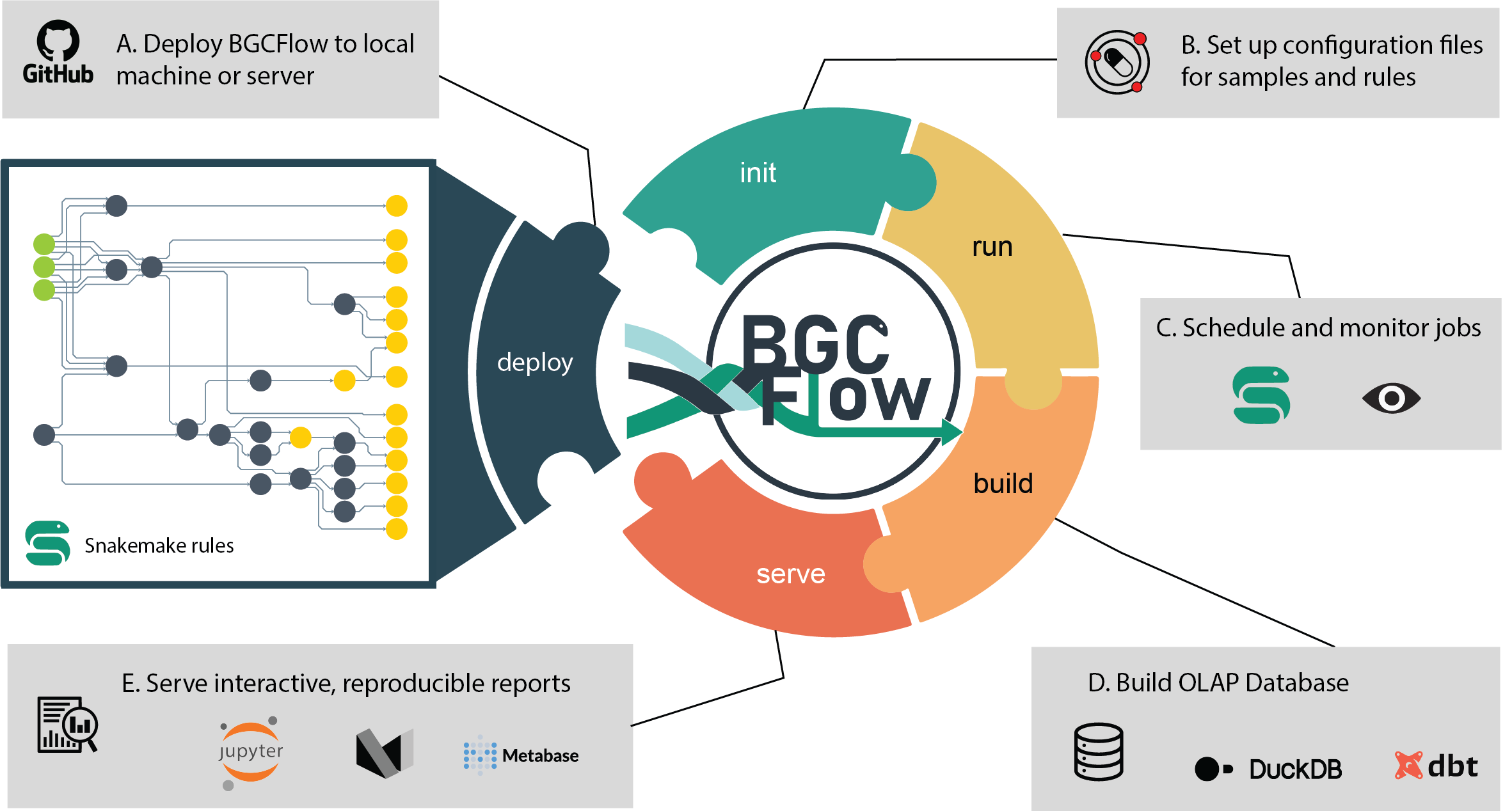
Features
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21 | $ bgcflow
Usage: bgcflow [OPTIONS] COMMAND [ARGS]...
A snakemake wrapper and utility tools for BGCFlow
(https://github.com/NBChub/bgcflow)
Options:
--version Show the version and exit.
-h, --help Show this message and exit.
Commands:
build Build Markdown report or use dbt to build DuckDB database.
clone Get a clone of BGCFlow to local directory.
deploy [EXPERIMENTAL] Deploy BGCFlow locally using snakedeploy.
get-result View a tree of a project results or get a copy using Rsync.
init Create projects or initiate BGCFlow config from template.
pipelines Get description of available pipelines from BGCFlow.
run A snakemake CLI wrapper to run BGCFlow.
serve Serve static HTML report or other utilities (Metabase, etc.).
sync Upload and sync DuckDB database to Metabase.
|
Credits
This package was created with the ppw tool. For more information, please visit the project page.
 ¶
¶

